p53-K382 monomethylated (R4BA1) Fab-His
From
$210.00
In stock
Only %1 left
SKU
2792
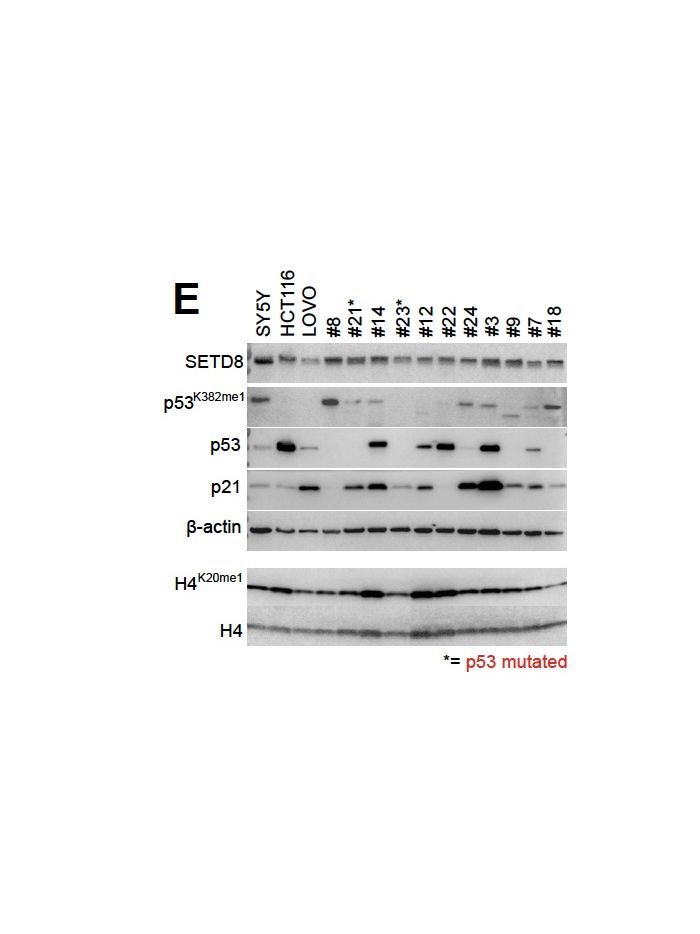
The lysine 382 residue (K382) in the C terminal regulatory domain of the p53 tumor suppressor is subject to monomethylation by the lysine methyltransferase SET8 (also known as PR Set7 or KMT5A) (1). This post-translational modification—p53 K382me1—is associated with suppression of p53’s transcriptional activation function. Methylated p53 at K382 shows reduced promoter binding to canonical target genes such as p21 (CDKN1A) and PUMA, both critical mediators of cell cycle arrest and apoptosis, thus restraining those pathways (2). Biochemically, K382me1 recruits chromatin compacting proteins like L3MBTL1, which further inhibits p53-driven gene expression at selected lociv (2). Notably, levels of K382 mono methylation decrease in response to DNA damage, indicating a dynamic regulatory switch during cellular stress (1,2).
Dysregulation of p53 K382me1 has implications in cancer biology. In tumor cells, elevated K382 methylation shifts p53’s transcriptional output away from apoptosis or cell-cycle arrest and towards DNA repair programs—for example, selective activation of GADD45 while repressing p21 and PUMA(2). This can permit NA-damaged cells to survive and potentially accumulate mutations, promoting tumor progression. In various cancers, overexpression or hyperactivity of SET8 and associated methyltransferases has been correlated with impaired p53 function and aggressive disease phenotypes(2,3). These insights support potential therapeutic strategies aimed at inhibiting K382 mono methylation—for example by targeting SET8—to reactivate p53-mediated tumor suppressive functions, enhance apoptosis, and improve DNA-damage responses in cancer treatments.